layout: true background-image: url(img/dlookr_hex.png) background-size: 80px background-position: 95% 95% --- name: xaringan-title class: left, middle background-image: url(img/white-abstract-background.jpg) background-size: cover <h1 style="color: gray;">Data diagnosis, exploration, and transformation with {dlookr}</h1> <img src="img/dlookr_hex.png" alt="dlookr-hex-sticker" width="180" /> <h3 style="color: gray;">Rachel Heise | WCM Computing Club | October 4, 2022</h3> --- # dlookr - R package for exploratory data analysis - Data diagnosis - Exploration - Transformation -- - What are the advantages of `dlookr`? - `dlookr` functions output dataframes for seamless integration with `dplyr` - Use of `plot` and `summary` functions to quickly view diagnostic data - Simple and powerful commands --- # Data .pull-left[ - Use `iris` dataset...with some NAs added for complexity - `iris` variables - Sepal Length (continuous, add NA) - Sepal Width (continuous) - Petal Length (continuous) - Petal Width (continuous) - Species (categorical, add NA) ] .pull-right[ ```r data("iris") set.seed(1) iris["Sepal.Length"] <- lapply(iris["Sepal.Length"], function(x) {x[sample( c(1:40), floor(40/10))] <- NA; x}) iris["Species"] <- lapply(iris["Species"], function(x) {x[sample( c(1:35), floor(35/10))] <- NA; x}) iris %>% data.table() ``` ``` Sepal.Length Sepal.Width Petal.Length Petal.Width Species 1: NA 3.5 1.4 0.2 setosa 2: 4.9 3.0 1.4 0.2 setosa 3: 4.7 3.2 1.3 0.2 setosa 4: NA 3.1 1.5 0.2 setosa 5: 5.0 3.6 1.4 0.2 setosa --- 146: 6.7 3.0 5.2 2.3 virginica 147: 6.3 2.5 5.0 1.9 virginica 148: 6.5 3.0 5.2 2.0 virginica 149: 6.2 3.4 5.4 2.3 virginica 150: 5.9 3.0 5.1 1.8 virginica ``` ] --- # Diagnose .pull-left[ - `diagnose` function describes: - the type of each variable - number & percent of missing data - unique values - Use `diagnose` for quick data diagnostics ] .pull-right[ ```r diagnose(iris) ``` ``` # A tibble: 5 × 6 variables types missing_count missing_percent unique_count unique_rate <chr> <chr> <int> <dbl> <int> <dbl> 1 Sepal.Length numeric 4 2.67 36 0.24 2 Sepal.Width numeric 0 0 23 0.153 3 Petal.Length numeric 0 0 43 0.287 4 Petal.Width numeric 0 0 22 0.147 5 Species factor 3 2 4 0.0267 ``` ] --- # Diagnose .pull-left[ - `diagnose` outputs a dataframe - This dataframe can be used with `dplyr` to perform any operations, including sorting and filtering ] -- .pull-right[ ```r iris %>% diagnose() %>% select(-unique_count, -unique_rate) %>% filter(missing_count > 0) %>% arrange(desc(missing_count)) ``` ``` # A tibble: 2 × 4 variables types missing_count missing_percent <chr> <chr> <int> <dbl> 1 Sepal.Length numeric 4 2.67 2 Species factor 3 2 ``` ] --- # Diagnose Continuous Variables .pull-left[ - `diagnose_numeric()` calculates min, max, mean, median, Q1, Q3, number of zeros, negative values, and outliers ```r diagnose_numeric(iris) ``` ``` # A tibble: 4 × 10 variables min Q1 mean median Q3 max zero minus outlier <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <int> <int> 1 Sepal.Length 4.3 5.1 5.87 5.8 6.4 7.9 0 0 0 2 Sepal.Width 2 2.8 3.06 3 3.3 4.4 0 0 4 3 Petal.Length 1 1.6 3.76 4.35 5.1 6.9 0 0 0 4 Petal.Width 0.1 0.3 1.20 1.3 1.8 2.5 0 0 0 ``` ] -- .pull-right[ - `diagnose_outlier()` identifies the mean of each variable with outliers, without outliers, and the mean of the outliers themselves ```r diagnose_outlier(iris) ``` ``` variables outliers_cnt outliers_ratio outliers_mean with_mean without_mean 1 Sepal.Length 0 0.000000 NaN 5.869178 5.869178 2 Sepal.Width 4 2.666667 3.675 3.057333 3.040411 3 Petal.Length 0 0.000000 NaN 3.758000 3.758000 4 Petal.Width 0 0.000000 NaN 1.199333 1.199333 ``` ```r diagnose_outlier(iris) %>% filter(outliers_cnt > 0) ``` ``` variables outliers_cnt outliers_ratio outliers_mean with_mean without_mean 1 Sepal.Width 4 2.666667 3.675 3.057333 3.040411 ``` ] --- # Diagnose Continuous Variables ```r iris %>% plot_outlier(Sepal.Width) ``` <!-- --> --- # Diagnose Categorical Variables .pull-left[ - `diagnose_category` is used to assess categorical variables - Levels of each categorical variable - Number & percentage of observations at each level - Rank of each level by number of observations ] .pull-right[ ```r diagnose_category(iris) ``` ``` # A tibble: 4 × 6 variables levels N freq ratio rank <chr> <chr> <int> <int> <dbl> <int> 1 Species versicolor 150 50 33.3 1 2 Species virginica 150 50 33.3 1 3 Species setosa 150 47 31.3 3 4 Species <NA> 150 3 2 4 ``` ] -- .pull-right[ ```r diagnose_category(iris) %>% filter(is.na(levels)) ``` ``` # A tibble: 1 × 6 variables levels N freq ratio rank <chr> <chr> <int> <int> <dbl> <int> 1 Species <NA> 150 3 2 4 ``` ] --- # Missing Values .pull-left[ - Use `plot_na_pareto()` to assess the highest quantity of missing values and `plot_na_hclust()` to assess the distribution of missing values ```r iris %>% plot_na_pareto(only_na = TRUE) ``` <!-- --> ] -- .pull-right[ ```r plot_na_hclust(iris) ``` <!-- --> ] --- # Describe .pull-left[ - Compute descriptive statistics for numerical data with `describe` ```r describe(iris) ``` ``` # A tibble: 4 × 26 described_varia… n na mean sd se_mean IQR skewness kurtosis p00 <chr> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> 1 Sepal.Length 146 4 5.87 0.821 0.0680 1.3 0.304 -0.551 4.3 2 Sepal.Width 150 0 3.06 0.436 0.0356 0.5 0.319 0.228 2 3 Petal.Length 150 0 3.76 1.77 0.144 3.5 -0.275 -1.40 1 4 Petal.Width 150 0 1.20 0.762 0.0622 1.5 -0.103 -1.34 0.1 # … with 16 more variables: p01 <dbl>, p05 <dbl>, p10 <dbl>, p20 <dbl>, # p25 <dbl>, p30 <dbl>, p40 <dbl>, p50 <dbl>, p60 <dbl>, p70 <dbl>, # p75 <dbl>, p80 <dbl>, p90 <dbl>, p95 <dbl>, p99 <dbl>, p100 <dbl> ``` ] -- .pull-right[ - Use with `group_by()` from `dplyr` to assess descriptive statistics by a categorical variable ```r iris %>% group_by(Species) %>% describe(Sepal.Length) ``` ``` # A tibble: 4 × 27 described_variables Species n na mean sd se_mean IQR skewness <chr> <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> 1 Sepal.Length setosa 43 4 5.04 0.330 0.0504 0.400 0.257 2 Sepal.Length versicolor 50 0 5.94 0.516 0.0730 0.7 0.105 3 Sepal.Length virginica 50 0 6.59 0.636 0.0899 0.675 0.118 4 Sepal.Length <NA> 3 0 4.67 0.404 0.233 0.400 0.722 # … with 18 more variables: kurtosis <dbl>, p00 <dbl>, p01 <dbl>, p05 <dbl>, # p10 <dbl>, p20 <dbl>, p25 <dbl>, p30 <dbl>, p40 <dbl>, p50 <dbl>, # p60 <dbl>, p70 <dbl>, p75 <dbl>, p80 <dbl>, p90 <dbl>, p95 <dbl>, # p99 <dbl>, p100 <dbl> ``` ] --- # Normality .pull-left[ - Assess normality of continuous variables and plot log and sqrt transformations with `normality` function - Shapiro-Wilk test is used to generate p-values - Plot the results with `plot_normality` to see the result of some basic transformations ```r normality(iris) ``` ``` # A tibble: 4 × 4 vars statistic p_value sample <chr> <dbl> <dbl> <dbl> 1 Sepal.Length 0.977 1.37e- 2 150 2 Sepal.Width 0.985 1.01e- 1 150 3 Petal.Length 0.876 7.41e-10 150 4 Petal.Width 0.902 1.68e- 8 150 ``` ] -- .pull-right[ ```r iris %>% plot_normality(Petal.Length) ``` <!-- --> ] --- # Correlation .pull-left[ - `correlate` function assesses the correlation between all continuous variables - Plotting the output of this function generates a correlation plot ```r correlate(iris) ``` ``` # A tibble: 12 × 3 var1 var2 coef_corr <fct> <fct> <dbl> 1 Sepal.Width Sepal.Length -0.109 2 Petal.Length Sepal.Length 0.870 3 Petal.Width Sepal.Length 0.814 4 Sepal.Length Sepal.Width -0.109 5 Petal.Length Sepal.Width -0.428 6 Petal.Width Sepal.Width -0.366 7 Sepal.Length Petal.Length 0.870 8 Sepal.Width Petal.Length -0.428 9 Petal.Width Petal.Length 0.963 10 Sepal.Length Petal.Width 0.814 11 Sepal.Width Petal.Width -0.366 12 Petal.Length Petal.Width 0.963 ``` ] -- .pull-right[ ```r iris %>% correlate() %>% plot() ``` <!-- --> ] --- # Target Variables .pull-left[ - Target variables are created using the `target_by()` function and are used to explore the relationship between a categorical variable and other variable values - Target variables are similar to `group_by()` in `dplyr` ] -- .pull-right[ ```r categ <- target_by(iris, Species) # Species is the target variable cat_num <- relate(categ, Petal.Length) # petal length is the numerical variable of interest plot(cat_num) ``` <!-- --> ] --- # Imputation - continuous .pull-left[ - Imputation methods for continuous variables include: - mean - median - mode - K-nearest neighbors (knn) - Recursive Partitioning and Regression Trees (rpart) - Multivariate Imputation by Chained Equations (mice) ] -- .pull-right[ ```r sepallength_im <- imputate_na(iris, Sepal.Length, yvar = Petal.Length, method = "knn") summary(sepallength_im) ``` ``` * Impute missing values based on K-Nearest Neighbors - method : knn * Information of Imputation (before vs after) Original Imputation described_variables "value" "value" n "146" "150" na "4" "0" mean "5.869178" "5.846767" sd "0.8211485" "0.8224163" se_mean "0.06795873" "0.06715001" IQR "1.3" "1.3" skewness "0.3041458" "0.3407754" kurtosis "-0.5505983" "-0.5545574" p00 "4.3" "4.3" p01 "4.4" "4.4" p05 "4.7" "4.7" p10 "4.900000" "4.801308" p20 "5" "5" p25 "5.1" "5.1" p30 "5.40" "5.27" p40 "5.6" "5.6" p50 "5.8" "5.8" p60 "6.1" "6.1" p70 "6.3" "6.3" p75 "6.4" "6.4" p80 "6.60" "6.52" p90 "6.9" "6.9" p95 "7.275" "7.255" p99 "7.7" "7.7" p100 "7.9" "7.9" ``` ] --- # Imputation - continuous .pull-left[ - Imputation methods for continuous variables include: - mean - median - mode - K-nearest neighbors (knn) - Recursive Partitioning and Regression Trees (rpart) - Multivariate Imputation by Chained Equations (mice) ] .pull-right[ ```r sepallength_im <- imputate_na(iris, Sepal.Length, yvar = Petal.Length, method = "knn") ``` ```r plot(sepallength_im) ``` <!-- --> ] --- # Imputation - categorical .pull-left[ - Imputation for categorical variables include: - mode - Recursive Partitioning and Regression Trees (rpart) - Multivariate Imputation by Chained Equations (mice) ```r species_im <- imputate_na(iris, Species, method = "rpart") ``` ```r summary(species_im) ``` ``` * Impute missing values based on Recursive Partitioning and Regression Trees - method : rpart * Information of Imputation (before vs after) original imputation original_percent imputation_percent setosa 47 50 31.33 33.33 versicolor 50 50 33.33 33.33 virginica 50 50 33.33 33.33 <NA> 3 0 2.00 0.00 ``` ] -- .pull-right[ ```r plot(species_im) ``` <!-- --> ] --- # Imputation - outliers .pull-left[ - Imputation methods for outliers: - mean - median - mode - capping (imputes upper outliers at 95th percentile and lower outliers at 5th percentile) ```r iris_outliers <- imputate_outlier(iris, Sepal.Width, method="capping") summary(iris_outliers) ``` ``` Impute outliers with capping * Information of Imputation (before vs after) Original Imputation described_variables "value" "value" n "150" "150" na "0" "0" mean "3.057333" "3.050967" sd "0.4358663" "0.4108379" se_mean "0.03558833" "0.03354478" IQR "0.5" "0.5" skewness "0.3189657" "0.1340705" kurtosis " 0.2282490" "-0.4114289" p00 "2.0" "2.2" p01 "2.2" "2.2" p05 "2.34500" "2.36975" p10 "2.5" "2.5" p20 "2.7" "2.7" p25 "2.8" "2.8" p30 "2.8" "2.8" p40 "3" "3" p50 "3" "3" p60 "3.1" "3.1" p70 "3.2" "3.2" p75 "3.3" "3.3" p80 "3.4" "3.4" p90 "3.61" "3.61" p95 "3.8" "3.8" p99 "4.151" "3.900" p100 "4.4" "4.0" ``` ] -- .pull-right[ ```r plot(iris_outliers) ``` 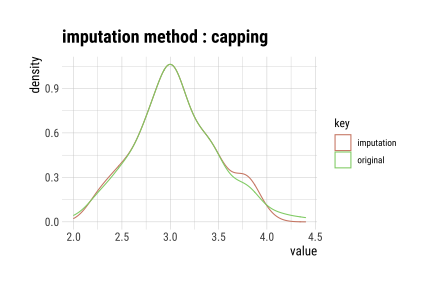<!-- --> ] --- # Standardization and Skewness .pull-left[ - `transform` function can both standardize (using z-score or minmax methods) and resolve skewness (via various transformations). ] -- .pull-right[ ```r iris_t <- transform(iris$Sepal.Length, method = "minmax") plot(iris_t) ``` 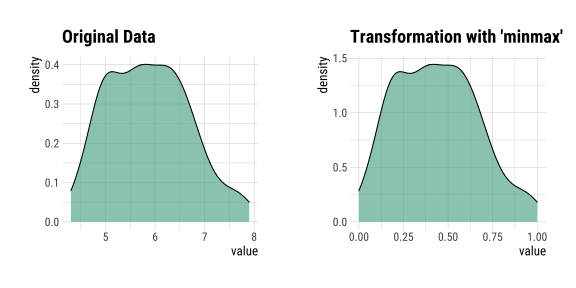<!-- --> ] --- # Find Skewness .pull-left[ - `find_skewness` function identifies skewed variables. Use `index=FALSE` argument to return the names of the skewed variables. ] -- .pull-right[ ```r find_skewness(iris, index = FALSE) ``` ``` character(0) ``` ```r find_skewness(iris, value = TRUE) ``` ``` Sepal.Length Sepal.Width Petal.Length Petal.Width 0.301 0.316 -0.272 -0.102 ``` ```r #filter on level of skewness find_skewness(iris, value = TRUE, thres = 0.3) ``` ``` Sepal.Length Sepal.Width 0.301 0.316 ``` ] --- # Binning .pull-left[ - `binning` transforms a continuous variable into a categorical variable. - It includes the following types of binning: - quantile - equal - pretty (Computes equally spaced ‘round’ values that are 1, 2 or 5 times a power of 10) - kmeans - bclust (bagged clustering) ] -- .pull-right[ ```r bin <- binning(iris$Petal.Length, nbins = 4, type = "kmeans", labels = c("B1", "B2", "B3", "B4")) #default type is quantile bin ``` ``` binned type: kmeans number of bins: 4 x B1 B2 B3 B4 50 25 45 30 ``` ] --- # Binning .pull-left[ ```r summary(bin) ``` ``` levels freq rate 1 B1 50 0.3333333 2 B2 25 0.1666667 3 B3 45 0.3000000 4 B4 30 0.2000000 ``` ] .pull-right[ ```r plot(bin) ``` 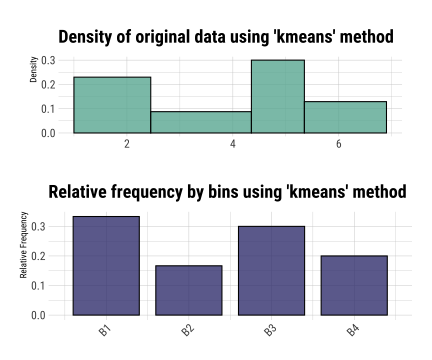<!-- --> ] --- # Questions? <style type="text/css"> .remark-code{line-height: 1.5; font-size: 60%} pre { max-width: 600px; max-height: 200px; overflow-x: auto; overflow-y: scroll; font-weight: 300; font-style: initial; } </style>